Eran Segal
Dean of the Biological and Life Sciences Division, and Professor of Computational Biology
Research Interests
Professor Segal’s research focuses on developing multi-modal AI models for personalized medicine based on big data from human cohorts, including microbiome, genetics, nutrition, and lifestyle data. Email

Prior to joining MBZUAI, Professor Segal published more than 200 peer-reviewed publications, which have been cited more than 60,000 times. He received several awards and honors for his work, including the Overton prize, awarded annually by the International Society for Bioinformatics (ICSB) to one scientist for outstanding accomplishments in computational biology, and the Michael Bruno award. He heads the Human Phenotype Project, a large-scale (more than 10,000 participants) deep-phenotype prospective longitudinal cohort and biobank that his lab established, aimed at identifying novel molecular markers with diagnostic, prognostic and therapeutic value, and at developing prediction models for disease onset and progression. The deep profiling includes medical history, lifestyle and nutritional habits, vital signs, anthropometrics, blood tests, continuous glucose and sleep monitoring, and molecular profiling of the transcriptome, genetics, gut and oral microbiome, metabolome and immune system.
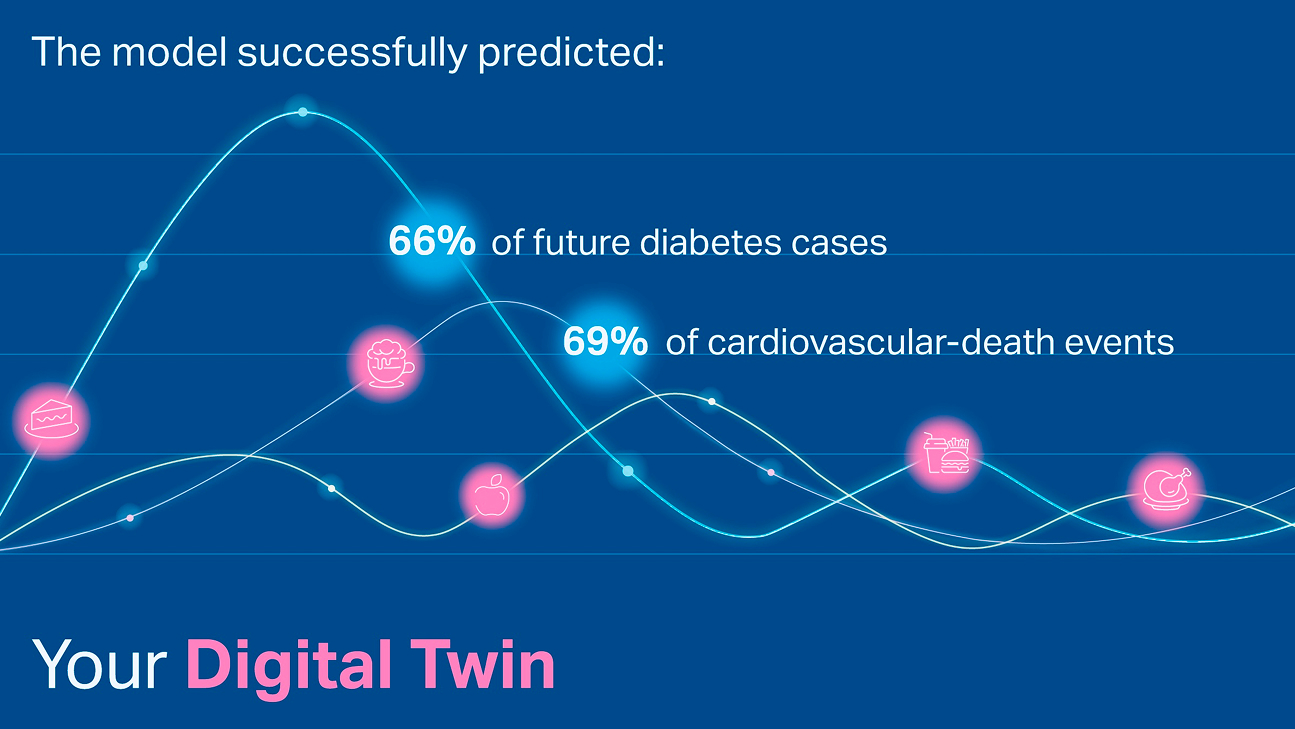
Segal’s analysis of this data provides novel insights into potential drivers of obesity, diabetes, and heart disease, and identifies hundreds of novel markers at the microbiome, metabolite, and immune system level. The predictive models developed can be translated into personalized disease prevention and treatment plans, and to the development of new therapeutic modalities based on metabolites and the microbiome.
Segal’s research focuses on developing and fine-tuning robust foundation models via Self-supervised Learning (SSL) techniques based on the data of the Human Phenotype Project. The models draw inspiration from large language models (LLMs) like OpenAI’s GPT-series models, but extend them by handling diverse clinical and multi-omics data modalities, including imaging, time-series, tabular, and sequencing-based data. While LLMs can understand and generate human-like text, here the focus is on creating multi-modal models that can effectively analyze heterogeneous types of medical data and be tailored to the biomedical domain.
The multi-modal models developed excel at extracting meaningful features and patterns from each data modality while capturing the complex interplay between different modalities. For example, when analyzing a patient's health status, our models can simultaneously consider their genetic information, imaging data, time-series data from wearables, and tabular data like blood tests. By learning the relationships between these modalities, the models can generate holistic insights into an individual's health, predict disease risks, stratify patients, and identify optimal treatment strategies. By combining different data modalities, the research can unlock new avenues for predictive modeling, disease diagnosis, and personalized medicine. This multi-modal approach has the potential to revolutionize healthcare by harnessing the power of AI to analyze and interpret complex medical data across different modalities.
Segal’s lab includes a multi-disciplinary team of computational biologists and experimental scientists in the area of computational and systems biology. The group has extensive experience in machine learning, computational biology, and analysis of heterogeneous high-throughput genomic data.
He was elected as an EMBO member and as a member of the young Israeli academy of science. During the COVID-19 pandemic, Professor Segal developed models for analyzing the dynamics of the pandemic and served as a senior advisor to the government of Israel.
Segal was awarded a B.Sc. in Computer Science summa cum laude from Tel-Aviv University, and a Ph.D. in Computer Science and Genetics, from Stanford University. Before joining the Weizmann Institute, Professor Segal held an independent research position at Rockefeller University, New York.
Segal’s analysis of this data provides novel insights into potential drivers of obesity, diabetes, and heart disease, and identifies hundreds of novel markers at the microbiome, metabolite, and immune system level. The predictive models developed can be translated into personalized disease prevention and treatment plans, and to the development of new therapeutic modalities based on metabolites and the microbiome.
Segal’s research focuses on developing and fine-tuning robust foundation models via Self-supervised Learning (SSL) techniques based on the data of the Human Phenotype Project. The models draw inspiration from large language models (LLMs) like OpenAI’s GPT-series models, but extend them by handling diverse clinical and multi-omics data modalities, including imaging, time-series, tabular, and sequencing-based data. While LLMs can understand and generate human-like text, here the focus is on creating multi-modal models that can effectively analyze heterogeneous types of medical data and be tailored to the biomedical domain.
The multi-modal models developed excel at extracting meaningful features and patterns from each data modality while capturing the complex interplay between different modalities. For example, when analyzing a patient's health status, our models can simultaneously consider their genetic information, imaging data, time-series data from wearables, and tabular data like blood tests. By learning the relationships between these modalities, the models can generate holistic insights into an individual's health, predict disease risks, stratify patients, and identify optimal treatment strategies. By combining different data modalities, the research can unlock new avenues for predictive modeling, disease diagnosis, and personalized medicine. This multi-modal approach has the potential to revolutionize healthcare by harnessing the power of AI to analyze and interpret complex medical data across different modalities.
Segal’s lab includes a multi-disciplinary team of computational biologists and experimental scientists in the area of computational and systems biology. The group has extensive experience in machine learning, computational biology, and analysis of heterogeneous high-throughput genomic data.
He was elected as an EMBO member and as a member of the young Israeli academy of science. During the COVID-19 pandemic, Professor Segal developed models for analyzing the dynamics of the pandemic and served as a senior advisor to the government of Israel.
Segal was awarded a B.Sc. in Computer Science summa cum laude from Tel-Aviv University, and a Ph.D. in Computer Science and Genetics, from Stanford University. Before joining the Weizmann Institute, Professor Segal held an independent research position at Rockefeller University, New York.
- Ph.D. in Computer Science, Stanford University, USA
- Ph.D. minor in genetics. Stanford University, USA
- B.Sc. in Computer Science, Tel-Aviv University, Israel
- 2023- Member of the Scientific Advisory Committee of the National AI Initiative of Israel
- 2020-1 Advisor to the government of Israel, COVID-19 modeling and combatting strategies
- Highly cited researcher, Web of Science, 2020
- Scripps Translational Science Award, 2019
- European Research Council (ERC) Advanced grant, 2018
- Elected to list of 50 Innovators, Sonima, 2016
- Michael Bruno prize, awarded by the Israel Institute for Advanced Studies, 2015
- Elected as EMBO Member, 2015
- European Research Council (ERC) Consolidator grant, 2014
- Landmark paper by the journal Cell (one of 25 papers in 1974-2014), 2014
- Third highest public engagement for a scientific paper - Almetric.com, 2014
- Elected to the Young Israeli Academy of Science, 2012
- Morris L. Levinson prize in biology, 2009
- Scientist to watch, The Scientist, 2008
- ESI-Topics hot paper, 2008
- European Research Council (ERC) starting grant (one of 201 proposals selected from more than 9000), 2008
- Overton prize, International Society for Computational Biology (ISCB), 2008 (awarded to one scientist a year, for outstanding accomplishments in computational biology)
- EMBO Young Investigator Award, 2007
- Alon Fellowship Tenure track for young faculty, Israel, 2006
- Research Fellowship, Center for Studies in Physics and Biology, Rockefeller University, 2004
- Best Paper by a Young Scientist Award, 2004 (Research in Computational Molecular Biology (RECOMB))
- Best Paper Award - Intelligent Systems for Molecular Biology (ISMB), 2003
- Best Student Paper Award - Intelligent Systems for Molecular Biology (ISMB), 2003
- Runner-up for Best Student Paper Award - Uncertainty in Artificial Intelligence (UAI), 2003
- Best Presentation Award - Biomedical Computation at Stanford (BCATS), 2003
- Stanford Graduate Fellowship (SGF), 1999-2003
- Fulbright Scholarship, 1999-2001
- Three-time Dean of the Faculty Award of Excellence, Faculty of exact sciences, Tel-Aviv University, 1995-1998
- Three-time Dean of the Faculty B.Sc. Scholarship, Faculty of exact sciences, Tel-Aviv University, 1995-1998
Segal has published more than 200 publications, and received several awards and honors for his work, including the Overton prize (awarded annually by the International Society for Bioinformatics (ICSB) to one scientist for outstanding accomplishments in computational biology).
- Talmor-Barkan et el., E. Segal. Metabolomic & microbiome profiling reveals personalized risk factors for coronary artery disease. Nature Medicine (2022), 28(2):303-314.
- Levin D, Raab N, et al., Pinto Y, et al., E. Segal. Diversity and functional landscapes in the microbiota of animals in the wild. Science (2021) Apr 16;372(6539):eabb5352.
- H. Rossman, S. Shilo, et al., U. Shalit, E. Segal. COVID-19 dynamics after a national immunization program in Israel. Nature Medicine (2021), 27(6):1055-1061.
- N. Bar, et al., E. Segal. A reference map of potential determinants for the human serum metabolome. Nature (2020), 588(7836):135-140.
- H. Rossman, A. Keshet, S. Shilo, A. Gavrieli, T. Bauman, O. Cohen, E. Shelly, R. Balicer, B. Geiger, Y. Dor, E. Segal. A framework for identifying regional outbreak and spread of COVID-19 from one-minute population-wide surveys. Nature Medicine (2020), 26(5):634-638.
- E. Blacher, et al., E. Segal*, E. Elinav*. Potential roles of gut microbiome & metabolites in modulation of murine ALS Nature (2019), 572(7770):474-480. * Senior authors
Contact faculty affairs
Interested in working with
our renowned faculty?
Fill out the below form and we will get back to you.