How MedNNS picks the right AI model for each type of hospital scan
Monday, September 29, 2025
If you’re using AI models to provide healthcare services today, you’re probably still spending weeks testing computer vision architectures and pretraining recipes for each new imaging task. Pick the wrong “model shape,” and the training can drag forever while accuracy plateaus; start from the wrong pretrained weights, and the network never really “locks on” to the right patterns. In medicine, where an X-ray looks nothing like a cat photo, defaulting to ImageNet (the classical image dataset used for model pretraining) is a shaky bet.
That’s the everyday pain point MedNNS targets. The system, to be presented at MICCAI 2025 in South Korea, promises to end the trial-and-error of building medical imaging AI by reframing model selection as a retrieval problem: given a new dataset, recommend the architecture and initialization most likely to win, before you spend GPU cycles. “Think of it as a matchmaker,” co-author and MBZUAI Ph.D. student Lotfi Mecharbat says. “You bring a dataset; it searches its space and returns the best candidates so you start training from a strong place.”
How MedNSS works
Medical images are different to everyday photos: grayscale radiographs, microscopic histology or OCT scans with layered textures don’t quite look like the selfie you took with your dog on a walk. As a result, features learned on everyday photos often don’t transfer to medical images, so the vaunted head start of ImageNet can be an illusion.
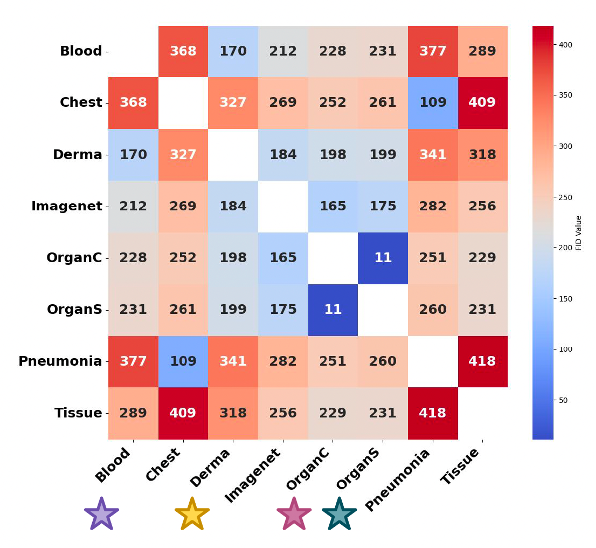
In the paper, the team quantifies this reality with Fréchet Inception Distance (FID): for each MedMNIST dataset, there’s usually another medical dataset that’s closer (lower FID) than ImageNet, evidence that domain-near pretraining should work better. They also show a simple proof-by-training-curve: pretraining on the closest medical dataset speeds up learning and improves the final score versus ImageNet.
Under the hood, MedNSS (short for Medical Neural Network Search) employs a Once-For-All (OFA) ResNet-like model, trained with a fairness-aware sampling schedule so rank among subnetworks is preserved under weight sharing.
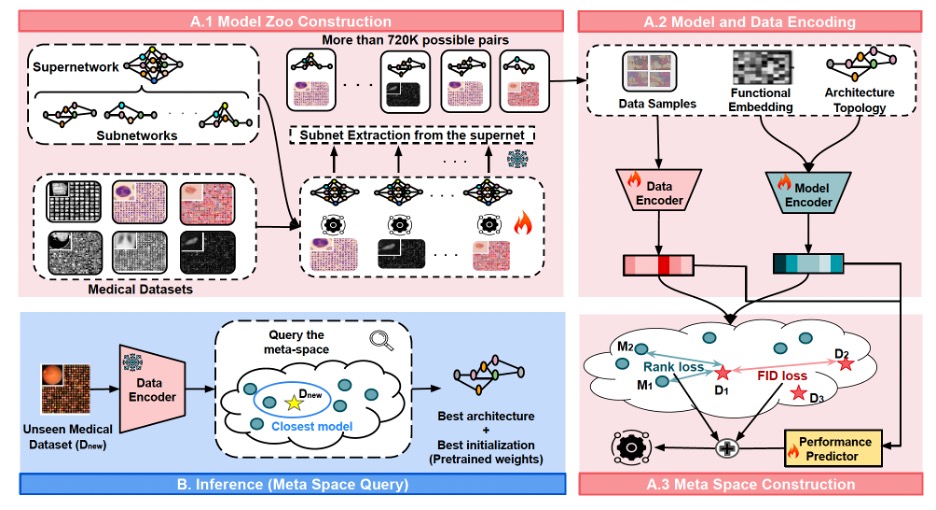
MedNNS is the first framework to the authors’ knowledge that jointly optimizes two decisions for a target medical dataset: the architecture and the pretrained weights. It builds a meta-space, a learned map where dataset embeddings and model embeddings live together. The scale of the model zoo is pretty significant: there are ~720k model–dataset pairs vs. ~14k in earlier task-adaptive search (≈51× larger). In that space, closeness predicts performance: models that sit near a dataset are likely to train faster and finish higher on it. At inference time, a new hospital dataset gets embedded and queried against this map to retrieve the best model(s).
MedNNS comes with three retrieval modes:
- T1: pick the single closest model and train it;
- T5 or T10: return a shortlist of 5 or 10, fine-tune them briefly (≈1 epoch), and continue with the best.
That shortlist trick balances speed and safety when the target task is unusual.
Previous “task-adaptive search” systems were computationally hungry because they trained huge numbers of model–dataset pairs to populate their lookup tables. MedNNS instead trains one Supernetwork per dataset, a large over-parameterized model (here, Once-For-All / OFA), and then extracts thousands of subnetworks by masking layers and channels, reusing weights via weight sharing. That yields a model zoo of ~720,000 model–dataset pairs (about 51× larger than prior work) without training 720,000 separate models. Crucially, the team trains the supernet in a way that preserves ranking among subnetworks, so a subnetwork that looks better during extraction really is better when trained from scratch. (They report ~90% Spearman rank correlation.)
“With 720k pairs instead of 14k, you get a far richer picture of what works where. The recommendations become reliable, not lucky,” according to Mecharbat.
The model embedding consists of two fingerprints combined: an architectural vector (depth, width, expansion ratios) plus a functional vector (how its penultimate features respond to a fixed input), passed through a small Multi-Layer Perceptron (MLP). The dataset embedding average features from a random batch of images, then another MLP.
A tiny performance predictor sits on top to learn how model–dataset pairs map to expected accuracy. But the real structure comes from two extra losses the authors add while training the meta-space: rank loss and FID loss. Rank loss enforces order: for a given dataset, higher-performing models must sit closer than weaker ones. FID loss pulls similar datasets together in the space, so experience transfers across related tasks.
Ablations show rank + FID together beat either alone or a contrastive baseline, creating a continuous space where “nearer is better” actually holds.
Where MedNSS works and where it does not
On the MedMNIST suite (Pneumonia, Organ, Tissue, Derm, Blood, Breast), MedNNS converges fast and finishes strong:
- Speed: At epoch 10, MedNNS often matches—or beats—competitors’ epoch-100 accuracy, a proxy for “time-to-useful-model.”
- Accuracy: On average, +1.7 percentage points over ImageNet-initialized baselines and SoTA NAS methods, with largest gains where domain-similar pretraining exists.
- Stability: T5/T10 shortlists give a small further bump while hedging bets on oddball datasets.
Clinically, the speed result may matter most: less GPU time means lower cost and quicker roll-outs when scanners change or screening programs spin up. “Hospitals don’t want to wait weeks,” Mecharbat said. “If you can hit a strong baseline in hours, the project survives.”
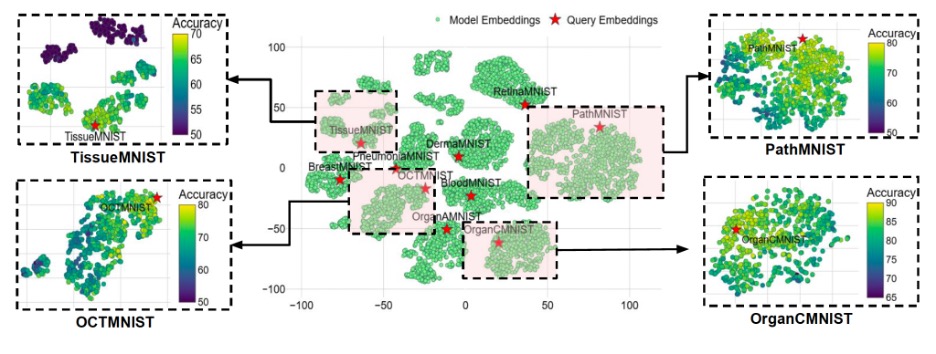
One dataset, TissueMNIST, is an outlier with a feature distribution far from others (high FID). There, similarity-based transfer helps less, and MedNNS’ edge shrinks. The lesson isn’t “don’t use MedNNS”; it’s “know when you’re off-distribution.” If a new dataset lands far from everything in the meta-space, the team recommends training from scratch or initializing from a larger medical foundation model.
Also: FID isn’t perfect. Designed for natural images, it can misread medical similarity (two histology sets may seem close yet be clinically distinct). The authors see room for domain-specific similarity measures by modality but warn that ultra-specialized metrics may not generalize across radiology, pathology, and ultrasound.
A hospital could actually use MedNSS today by uploading a small sample of their dataset and get its embedding. Then, they would query the meta-space and receive the T1/T5/T10 recommendations with pretrained weights. Finally, they’d fine-tune the chosen model(s); deploy the winner. For regional hospitals with small GPU budgets, the payoff is skipping dozens of dead-end trials. National programs could even host a shared meta-space so sites across the country get consistent, reproducible picks—useful for audits and multi-site studies.
What’s next: from images to everything, and from accuracy to constraints
The MBZUAI researchers want to take MedNSS into three directions:
- Beyond classification: Extend supernets and encoders to segmentation and detection, where architecture choice matters even more.
- Hardware-aware retrieval: Tag every candidate with VRAM/latency/energy so a clinic can ask, “best model under 8 GB VRAM?” and get filtered, ranked results.
- Foundation-model era: Fold self-supervised medical pretraining and foundation models into the model zoo, then adapt them task-by-task through the same meta-space logic. (The team says this is already underway.)
Longer term, Mecharbat imagines a living, community meta-space, governed by vendors, regulators, and hospitals, that updates as new datasets arrive, audits bias across demographics and scanners, and reports standardized metrics for transparency.
MedNNS doesn’t replace careful validation, but it replaces guesswork with guidance. By turning model selection into retrieval across a richly structured space, shaped by rank and data similarity, it gives medical AI teams a faster, more reliable path from “we have a dataset” to “we have a model that works.”
- healthcare ,
- conference ,
- MICCAI ,
- medical imaging ,
- images ,
Related
AI and the silver screen: how cinema has imagined intelligent machines
Movies have given audiences countless visions of how artificial intelligence might affect our lives. Here are some.....
- cinema ,
- AI ,
- artificial intelligence ,
- art ,
- fiction ,
- science fiction ,
Mind meld: agentic communication through thoughts instead of words
A NeurIPS 2025 study by MBZUAI shows that tapping into agents’ internal structures dramatically improves multi-agent decision-making.
- agents ,
- neurips ,
- machine learning ,
Balancing the future of AI: MBZUAI hosts AI for the Global South workshop
AI4GS brings together diverse voices from across continents to define the challenges that will guide inclusive AI.....
- representation ,
- equitable ,
- global south ,
- AI4GS ,
- event ,
- workshop ,
- languages ,
- inclusion ,
- large language models ,
- llms ,
- accessibility ,


